In this lab we will learn how to run and use continuum dielectric electrostatic calculations
using CHARMM through the MMTSB Tool Set.
1. Calculation of electrostatic solvation energies
We will use the engrailed homeodomain again. The structure is available from
the
Protein Data Bank
with the PDB code
1ENH.
First we need to obtain a structure
that has all of the atoms present (including hydrogens) and center the molecule
with respect to the origin:
convpdb.pl -nsel protein 1ENH.pdb > 1enh.protein.pdb
complete.pl 1enh.protein.pdb > 1enh.allh.pdb
convpdb.pl -center 1enh.allh.pdb > 1enh.center.pdb
Electrostatic solvation energies according to Poisson theory can be obtained
using the CHARMM PBEQ module with the following command:
pbCHARMM.pl 1enh.center.pdb
The Poisson equation is solved on a grid where the grid resolution determines
the accuracy of the result. You can change the grid resolution with the
parameter 'dcel':
pbCHARMM.pl -par dcel=0.5 1enh.center.pdb
Try using values from 0.5 (default) to 0.2. How does the
energy change?
The GBMV generalized Born approximation can also be used
to calculate electrostatic solvation energies. Try calculating the GB
energy for the same system with the following command:
enerCHARMM.pl -par gb,nocut -out gb 1enh.center.pdb
How much does the result deviate?
An alternative to the commonly employed molecular surface definition (that is
used by default) a smoother surface based on overlapping van der Waals spheres
is sometimes used. This surface is turned on with the 'smooth' parameter:
pbCHARMM.pl -par dcel=0.5,smooth 1enh.center.pdb
The GBSW method matches the smooth surface. GBSW energies are calculated with:
enerCHARMM.pl -par gb=gbsw,nocut -out gb 1enh.center.pdb
Compare all of the results obtained so far.
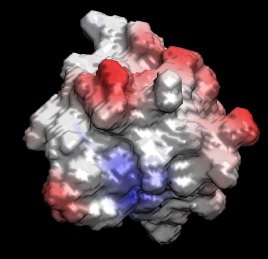
2. Calculation of electrostatic maps by solving the Poisson equation
Maps of electrostatic potentials around biomolecules are useful in determining possible
interaction sites for protein-protein and protein-ligand interactions. Electrostatic
potentials can be obtained by solving the Poisson equation for a given molecular structure.
In this exercise we will use CHARMM to carry out such a calculation and then use VMD
in order to visualize the results.
We will use the tool
pbCHARMM.pl again in order to calculate the electrostatic
potential on a grid:
pbCHARMM.pl -emap phi.dx 1enh.center.pdb
The electrostatic potential is best visualized by projection onto the molecular surface.
The molecular surface can be obtained from the distribution of the dielectric constant with:
pbCHARMM.pl -dx -grid epsx grid.dx 1enh.center.pdb
Now use VMD and load the molecule, the molecular surface, and the electrostatic potential.
Color the molecular surface according to the potential and compare with the location
of the charged residues on the surface.
Calculate the electrostatic map also for the smooth van der Waals-based surface and compare.
Because the surface has changed, both the electrostatic potential and surface need to
be recalculated.
You can change the dielectric constant and introduce a salt concentration of the solute.
The corresponding parameters are epsw (external dielectric constant) and pbionconc
(ion concentration). For example, epsilon=50 and an ion concentration of 0.1 M is modeled
with:
pbCHARMM.pl -emap modphi.dx -par epsw=50,pbionconc=0.1 1enh.center.pdb
Compare the resulting map with the result obtained previously.



